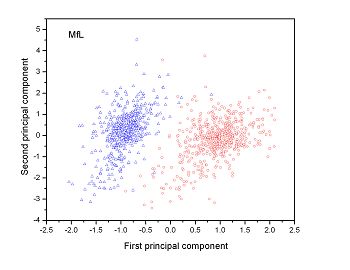
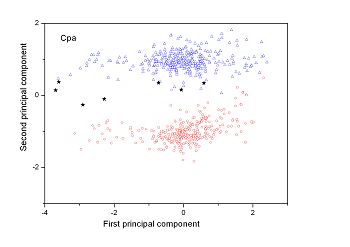
Figure 2 (a-n). The distribution of points on the principal plane spanned by the first (x) and second (y) principal axes using the principal component analysis in the 28 plant pathogens. The red circles represent the function-known genes, the blue triangles represent the corresponding negative samples and the black stars denote the ORFs recognized as non-coding. Note that in most figures, the distribution of the red circles is well separated from that of the blue triangles, indicating that coding and non-coding sequences are well distinguished. Furthermore, most of the identified non-coding ORFs distribute far from the core of red circles, and close to the core of blue triangles, implying that most of the recognized non-coding ORFs listed in Table 4 are unlikely to encode protein.
 |
 |
| (1) Acidovorax avenae subsp. citrulli AAC00-1 | (2) Agrobacterium tumefaciens str. C58 |
 |
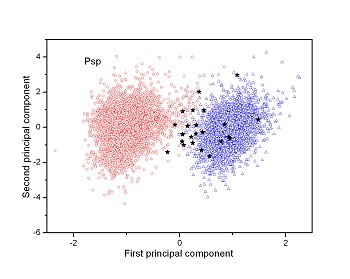 |
| (3) Erwinia carotovora subsp. atroseptica SCRI1043 | (4) Pseudomonas syringae pv. phaseolicola 1448A |
 |
 |
| (5) Pseudomonas syringae pv. syringae B728a | (6) Pseudomonas syringae pv. tomato str.DC3000 |
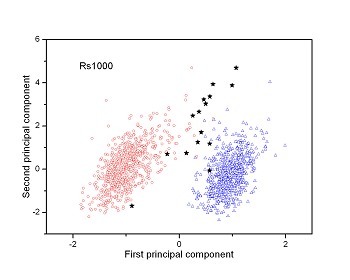 |
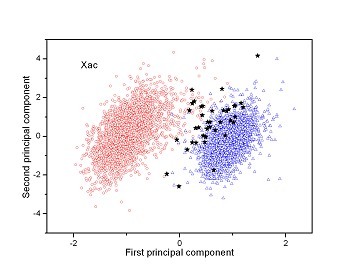 |
| (7) Ralstonia solanacearum GMI1000 | (8) Xanthomonas axonopodis pv. citri str. 306 |
 |
 |
| (9) Xanthomonas campestris pv. campestris str. 8004 | (10) Xanthomonas campestris pv. campestris str. ATCC 33913 |
 |
 |
| (11) Xanthomonas oryzae pv. oryzae KACC10331 | (l2) Xanthomonas campestris pv. vesicatoria str. 85-10 |
 |
 |
| (13) Xylella fastidiosa Temecula1 | (14) Xanthomonas oryzae pv. oryzae MAFF 311018 |
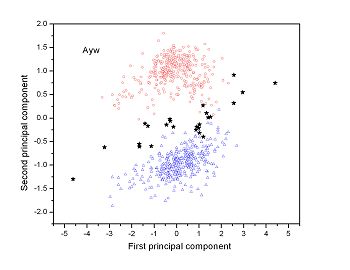 |
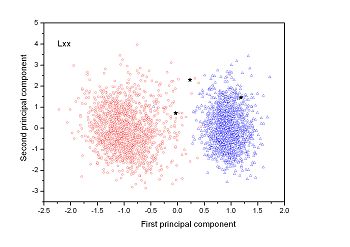 |
| (15) Aster yellows witches'-broom phytoplasma strain AY-WB | (16) Leifsonia xyli subsp. xyli str. CTCB07 |
 |
 |
| (17) Mesoplasma florum L1 | (18) Onion yellows phytoplasma OY-M |
 |
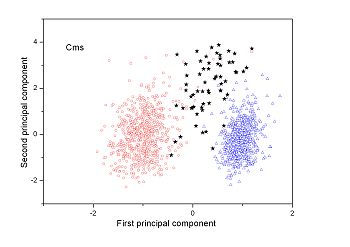 |
| (19)Agrobacterium vitis S4 | (20) Clavibacter michiganensis subsp. sepedonicus ATCC 33113 |
 |
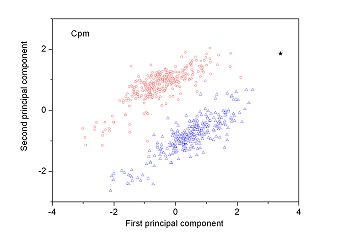 |
| (21) Candidatus Phytoplasma australiense | (22) Candidatus Phytoplasma mali |
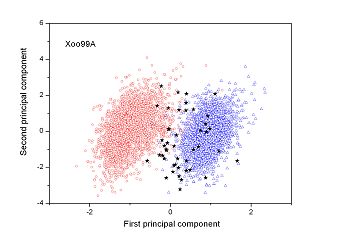 |
|
| (23) Xanthomonas oryzae pv. oryzae PXO99A |